Dask Dataframes#
Dask is a flexible parallel computing library for analytic computing written in Python. Dask is similar to Spark, by lazily constructing directed acyclic graph (DAG) of tasks and splitting large datasets into small portions called partitions. See the below image from Dask’s web page for illustration.
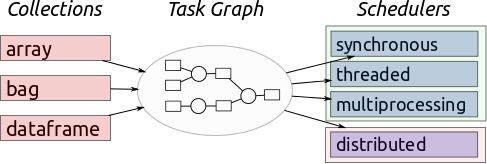
It has three main interfaces:
Bag, which is similar to RDD interface in Spark;
While it can work on a distributed cluster, Dask works also very well on a single cpu machine.
DataFrames#
Dask dataframes look and feel (mostly) like Pandas dataframes but they run on the same infrastructure that powers dask.delayed.
The dask.dataframe module implements a blocked parallel DataFrame object that mimics a large subset of the Pandas DataFrame. One dask DataFrame is comprised of many in-memory pandas DataFrames separated along the index. One operation on a dask DataFrame triggers many pandas operations on the constituent pandas DataFrames in a way that is mindful of potential parallelism and memory constraints.
Related Documentation
In this notebook, we will extracts some historical flight data for flights out of NYC between 1990 and 2000. The data is taken from here. This should only take a few seconds to run.
We will use dask.dataframe construct our computations for us. The dask.dataframe.read_csv function can take a globstring like "data/nycflights/*.csv" and build parallel computations on all of our data at once.
Variable descriptions
Name Description
Year1987-2008Month1-12DayofMonth1-31DayOfWee1 (Monday) - 7 (Sunday)DepTimeactual departure time (local, hhmm)CRSDepTimescheduled departure time (local, hhmm)ArrTimeactual arrival time (local, hhmm)CRSArrTimescheduled arrival time (local, hhmm)UniqueCarrierunique carrier codeFlightNumflight numberTailNuplane tail numberActualElapsedTimein minutesCRSElapsedTimein minutesAirTimein minutesArrDelayarrival delay, in minutesDepDelaydeparture delay, in minutesOriginorigin IATA airport codeDestdestination IATA airport codeDistancein milesTaxiIntaxi in time, in minutesTaxiOuttaxi out time in minutesCancelledwas the flight cancelled?CancellationCodereason for cancellation (A = carrier, B = weather, C = NAS, D = security)Diverted1 = yes, 0 = noCarrierDelayin minutesWeatherDelayin minutesNASDelayin minutesSecurityDelayin minutesLateAircraftDelayin minutes
Prep the Data#
import os
import pandas as pd
pd.set_option("max.rows", 10)
os.getcwd()
import os # library to get directory and file paths
import tarfile # this module makes possible to read and write tar archives
def extract_flight():
here = os.getcwd()
flightdir = os.path.join(here,'data', 'nycflights')
if not os.path.exists(flightdir):
print("Extracting flight data")
tar_path = os.path.join('data', 'nycflights.tar.gz')
with tarfile.open(tar_path, mode='r:gz') as flights:
flights.extractall('data/')
extract_flight() # this function call will extract 10 csv files in data/nycflights
Load Data from CSVs in Dask Dataframes#
import os
here = os.getcwd()
filenames = os.path.join(here, 'data', 'nycflights', '*.csv')
filenames
import dask
import dask.dataframe as dd
df = dd.read_csv(filenames,
parse_dates={'Date': [0, 1, 2]})
Let’s take a look to the dataframe
df
### Get the first 5 rows
df.head(5)
import traceback # we use traceback because we expect an error.
try:
df.tail(5) # Get the last 5 rows
except Exception:
traceback.print_exc()
What just happened?#
Unlike pandas.read_csv which reads in the entire file before inferring datatypes, dask.dataframe.read_csv only reads in a sample from the beginning of the file (or first file if using a glob). These inferred datatypes are then enforced when reading all partitions.
In this case, the datatypes inferred in the sample are incorrect. The first n rows have no value for CRSElapsedTime (which pandas infers as a float), and later on turn out to be strings (object dtype). When this happens you have a few options:
Specify dtypes directly using the
dtypekeyword. This is the recommended solution, as it’s the least error prone (better to be explicit than implicit) and also the most performant.Increase the size of the
samplekeyword (in bytes)Use
assume_missingto makedaskassume that columns inferred to beint(which don’t allow missing values) are actually floats (which do allow missing values). In our particular case this doesn’t apply.
In our case we’ll use the first option and directly specify the dtypes of the offending columns.
df.dtypes
df = dd.read_csv(filenames,
parse_dates={'Date': [0, 1, 2]},
dtype={'TailNum': object,
'CRSElapsedTime': float,
'Cancelled': bool})
df.tail(5)
Let’s take a look at one more example to fix ideas.
len(df)
Why df is ten times longer ?#
Dask investigated the input path and found that there are ten matching files.
A set of jobs was intelligently created for each chunk - one per original CSV file in this case.
Each file was loaded into a pandas dataframe, had
len()applied to it.The subtotals were combined to give you the final grant total.
Computations with dask.dataframe#
We compute the maximum of the DepDelay column. With dask.delayed we could create this computation as follows:
maxes = []
for fn in filenames:
df = dask.delayed(pd.read_csv)(fn)
maxes.append(df.DepDelay.max())
final_max = dask.delayed(max)(maxes)
final_max.compute()
Now we just use the normal Pandas syntax as follows:
%time df.DepDelay.max().compute()
This writes the delayed computation for us and then runs it. Recall that the delayed computation is a dask graph made of up of key-value pairs.
Some things to note:
As with
dask.delayed, we need to call.compute()when we’re done. Up until this point everything is lazy.Dask will delete intermediate results (like the full pandas dataframe for each file) as soon as possible.
This lets us handle datasets that are larger than memory
This means that repeated computations will have to load all of the data in each time (run the code above again, is it faster or slower than you would expect?)
As with Delayed objects, you can view the underlying task graph using the .visualize method:
df.DepDelay.max().visualize()
If you are already familiar with the Pandas API then know how to use dask.dataframe. There are a couple of small changes.
As noted above, computations on dask DataFrame objects don’t perform work, instead they build up a dask graph. We can evaluate this dask graph at any time using the .compute() method.
result = df.DepDelay.mean() # create the tasks graph
%time result.compute() # perform actual computation
Store Data in Apache Parquet Format#
Dask encourage dataframe users to store and load data using Parquet instead of csv. Apache Parquet is a columnar binary format that is easy to split into multiple files (easier for parallel loading) and is generally much simpler to deal with than HDF5 (from the Dask library’s perspective). It is also a common format used by other big data systems like Apache Spark and Apache Impala and so is useful to interchange with other systems.
df.drop("TailNum", axis=1).to_parquet("nycflights/") # save csv files using parquet format
It is possible to specify dtypes and compression when converting. This can definitely help give you significantly greater speedups, but just using the default settings will still be a large improvement.
df.size.compute()
import dask.dataframe as dd
df = dd.read_parquet("nycflights/")
df.head()
result = df.DepDelay.mean()
%time result.compute()
The computation is much faster because pulling out the DepDelay column is easy for Parquet.
Parquet advantages:#
Binary representation of data, allowing for speedy conversion of bytes-on-disk to bytes-in-memory
Columnar storage, meaning that you can load in as few columns as you need without loading the entire dataset
Row-chunked storage so that you can pull out data from a particular range without touching the others
Per-chunk statistics so that you can find subsets quickly
Compression
Exercise 15.1#
If you don’t remember how to use pandas. Please read pandas documentation.
Use the
head()method to get the first ten rowsHow many rows are in our dataset?
Use selections
df[...]to find how many positive (late) and negative (early) departure times there areIn total, how many non-cancelled flights were taken? (To invert a boolean pandas Series s, use ~s).
Divisions and the Index#
The Pandas index associates a value to each record/row of your data. Operations that align with the index, like loc can be a bit faster as a result.
In dask.dataframe this index becomes even more important. Recall that one dask DataFrame consists of several Pandas DataFrames. These dataframes are separated along the index by value. For example, when working with time series we may partition our large dataset by month.
Recall that these many partitions of our data may not all live in memory at the same time, instead they might live on disk; we simply have tasks that can materialize these pandas DataFrames on demand.
Partitioning your data can greatly improve efficiency. Operations like loc, groupby, and merge/join along the index are much more efficient than operations along other columns. You can see how your dataset is partitioned with the .divisions attribute. Note that data that comes out of simple data sources like CSV files aren’t intelligently indexed by default. In these cases the values for .divisions will be None.
df = dd.read_csv(filenames,
dtype={'TailNum': str,
'CRSElapsedTime': float,
'Cancelled': bool})
df.divisions
However if we set the index to some new column then dask will divide our data roughly evenly along that column and create new divisions for us. Warning, set_index triggers immediate computation.
df2 = df.set_index('Year')
df2.divisions
We see here the minimum and maximum values (1990 and 1999) as well as the intermediate values that separate our data well. This dataset has ten partitions, as the final value is assumed to be the inclusive right-side for the last bin.
df2.npartitions
df2.head()
One of the benefits of this is that operations like loc only need to load the relevant partitions
df2.loc[1991]
df2.loc[1991].compute()
Exercises 15.2#
In this section we do a few dask.dataframe computations. If you are comfortable with Pandas then these should be familiar. You will have to think about when to call compute.
In total, how many non-cancelled flights were taken from each airport?
Hint: use df.groupby. df.groupby(df.A).B.func().
What was the average departure delay from each airport?
Note, this is the same computation you did in the previous notebook (is this approach faster or slower?)
What day of the week has the worst average departure delay?


