F2py#
F2py is a part of Numpy and there are three ways to wrap Fortran with Python :
Write some fortran subroutines and just run f2py to create Python modules.
Insert special f2py directives inside Fortran source for complex wrapping.
Write a interface file (.pyf) to wrap Fortran files without changing them. f2py automatically generate the pyf template file that can be modified.
import os, sys
import matplotlib.pyplot as plt
import scipy.fftpack as sf
import scipy.linalg as sl
import numpy as np
Simple Fortran subroutine to compute norm#
Fortran 90/95 free format#
%%file euclidian_norm.f90
subroutine euclidian_norm (a, b, c)
real(8), intent(in) :: a, b
real(8), intent(out) :: c
c = sqrt (a*a+b*b)
end subroutine euclidian_norm
Writing euclidian_norm.f90
Fortran 77 fixed format#
%%file euclidian_norm.f
subroutine euclidian_norm (a, b, c)
real*8 a,b,c
Cf2py intent(out) c
c = sqrt (a*a+b*b)
end
Writing euclidian_norm.f
Build extension module with f2py program#
!{sys.executable} -m numpy.f2py --quiet -c euclidian_norm.f90 -m vect
/usr/share/miniconda/envs/python-fortran/lib/python3.9/site-packages/numpy/f2py/f2py2e.py:723: VisibleDeprecationWarning:
distutils has been deprecated since NumPy 1.26.x
Use the Meson backend instead, or generate wrappers without -c and use a custom build script
builder = build_backend(
/usr/share/miniconda/envs/python-fortran/lib/python3.9/site-packages/setuptools/_distutils/cmd.py:66: SetuptoolsDeprecationWarning: setup.py install is deprecated.
!!
********************************************************************************
Please avoid running ``setup.py`` directly.
Instead, use pypa/build, pypa/installer or other
standards-based tools.
See https://blog.ganssle.io/articles/2021/10/setup-py-deprecated.html for details.
********************************************************************************
!!
self.initialize_options()
WARN: Could not locate executable armflang
Removing build directory /tmp/tmpjhvb6qg0
Use the extension module in Python#
import vect
c = vect.euclidian_norm(3,4)
c
5.0
print(vect.euclidian_norm.__doc__) # Docstring is automatically generate
c = euclidian_norm(a,b)
Wrapper for ``euclidian_norm``.
Parameters
----------
a : input float
b : input float
Returns
-------
c : float
Fortran magic#
Jupyter extension that help to use fortran code in an interactive session.
It adds a %%fortran cell magic that compile and import the Fortran code in the cell, using F2py.
The contents of the cell are written to a .f90 file in the directory IPYTHONDIR/fortran using a filename with the hash of the code. This file is then compiled. The resulting module is imported and all of its symbols are injected into the user’s namespace.
%load_ext fortranmagic
F2py directives#
F2PY introduces also some extensions to Fortran 90/95 language specification that help designing Fortran to Python interface, make it more “Pythonic”.
If editing Fortran codes is acceptable, these specific attributes can be inserted directly to Fortran source codes. Special comment lines are ignored by Fortran compilers but F2PY interprets them as normal lines.
%%fortran
subroutine euclidian_norm(a,c,n)
integer :: n
real(8),dimension(n),intent(in) :: a
!f2py optional , depend(a) :: n=len(a)
real(8),intent(out) :: c
real(8) :: sommec
integer :: i
sommec = 0
do i=1,n
sommec=sommec+a( i )*a( i )
end do
c = sqrt (sommec)
end subroutine euclidian_norm
a=[2,3,4] # Python list
type(a)
list
euclidian_norm(a)
5.385164807134504
a=np.arange(2,5) # numpy array
type(a)
numpy.ndarray
euclidian_norm(a)
5.385164807134504
print(euclidian_norm.__doc__) # Documentation
c = euclidian_norm(a,[n])
Wrapper for ``euclidian_norm``.
Parameters
----------
a : input rank-1 array('d') with bounds (n)
Other Parameters
----------------
n : input int, optional
Default: shape(a, 0)
Returns
-------
c : float
directives#
optional: The corresponding argument is moved to the end.required: This is default. Use it to disable automatic optional setting.intent(in | inout | out | hide),intent(in)is the default.intent(out)is implicitly translated tointent(out,hide).intent(copy)andintent(overwrite)control changes for input arguments.checkperforms some assertions, it is often automatically generated.depend: f2py detects cyclic dependencies.allocatable, parameterintent(callback), external: for function as arguments.intent(c)C-type argument , array or function.C expressions:
rank, shape, len, size, slen.
Callback#
You can call a python function inside your fortran code
%%fortran
subroutine sum_f (f ,n, s)
!Compute sum(f(i), i=1,n)
external f
integer, intent(in) :: n
real, intent(out) :: s
s = 0.0
do i=1,n
s=s+f(i)
end do
end subroutine sum_f
def fonction(i) : # python function
return i*i
sum_f(fonction,3)
14.0
sum_f(lambda x :x**2,3) # lambda function
14.0
Fortran arrays and Numpy arrays#
Let’s see how to pass numpy arrays to fortran subroutine.
%%fortran --extra="-DF2PY_REPORT_ON_ARRAY_COPY=1"
subroutine push( positions, velocities, dt, n)
integer, intent(in) :: n
real(8), intent(in) :: dt
real(8), dimension(n,3), intent(in) :: velocities
real(8), dimension(n,3) :: positions
do i = 1, n
positions(i,:) = positions(i,:) + dt*velocities(i,:)
end do
end subroutine push
positions = [[0, 0, 0], [0, 0, 0], [0, 0, 0]]
velocities = [[0, 1, 2], [0, 3, 2], [0, 1, 3]]
import sys
push(positions, velocities, 0.1)
positions # memory is not updated because we used C memory storage
created an array from object
created an array from object
[[0, 0, 0], [0, 0, 0], [0, 0, 0]]
During execution, the message “created an array from object” is displayed, because a copy of is made when passing multidimensional array to fortran subroutine.
positions = np.array(positions, dtype='f8', order='F')
push(positions, velocities, 0.1)
positions # the memory is updated
created an array from object
array([[0. , 0.1, 0.2],
[0. , 0.3, 0.2],
[0. , 0.1, 0.3]])
Signature file#
This file contains descriptions of wrappers to Fortran or C functions, also called as signatures of the functions. F2PY can create initial signature file by scanning Fortran source codes and catching all relevant information needed to create wrapper functions.
f2py vector.f90 -h vector.pyf
vector.pyf
! -*- f90 -*-
! Note: the context of this file is case sensitive.
subroutine euclidian_norm(a,c,n) ! in vector.f90
real(kind=8) dimension(n),intent(in) :: a
real(kind=8) intent(out) :: c
integer optional,check(len(a)>=n),depend(a) :: n=len(a)
end subroutine euclidian_norm
! This file was auto-generated with f2py (version:2).
! See http://cens.ioc.ee/projects/f2py2e/
Wrap lapack function dgemm with f2py#
Generate the signature file
!{sys.executable} -m numpy.f2py --quiet -m mylapack --overwrite-signature -h dgemm.pyf dgemm.f
rmbadname1: Replacing "max" with "max_bn".
! -*- f90 -*-
! Note: the context of this file is case sensitive.
python module mylapack ! in
interface ! in :mylapack
subroutine dgemm(transa,transb,m,n,k,alpha,a,lda,b,ldb,beta,c,ldc) ! in :mylapack:dgemm.f
character*1 :: transa
character*1 :: transb
integer :: m
integer :: n
integer :: k
double precision :: alpha
double precision dimension(lda,*) :: a
integer, optional,check(shape(a,0)==lda),depend(a) :: lda=shape(a,0)
double precision dimension(ldb,*) :: b
integer, optional,check(shape(b,0)==ldb),depend(b) :: ldb=shape(b,0)
double precision :: beta
double precision dimension(ldc,*) :: c
integer, optional,check(shape(c,0)==ldc),depend(c) :: ldc=shape(c,0)
end subroutine dgemm
end interface
end python module mylapack
! This file was auto-generated with f2py (version:2).
! See http://cens.ioc.ee/projects/f2py2e/
!{sys.executable} -m numpy.f2py --quiet -c dgemm.pyf -llapack -m mylapack
/usr/share/miniconda/envs/python-fortran/lib/python3.9/site-packages/numpy/f2py/f2py2e.py:723: VisibleDeprecationWarning:
distutils has been deprecated since NumPy 1.26.x
Use the Meson backend instead, or generate wrappers without -c and use a custom build script
builder = build_backend(
/usr/share/miniconda/envs/python-fortran/lib/python3.9/site-packages/setuptools/_distutils/cmd.py:66: SetuptoolsDeprecationWarning: setup.py install is deprecated.
!!
********************************************************************************
Please avoid running ``setup.py`` directly.
Instead, use pypa/build, pypa/installer or other
standards-based tools.
See https://blog.ganssle.io/articles/2021/10/setup-py-deprecated.html for details.
********************************************************************************
!!
self.initialize_options()
WARN: Could not locate executable armflang
Removing build directory /tmp/tmpv_6kmg7d
import numpy as np
import mylapack
a = np.array([[7,8],[3,4],[1,2]])
b = np.array([[1,2,3],[4,5,6]])
print("a=",a)
print("b=",b)
assert a.shape[1] == b.shape[0]
c = np.zeros((a.shape[0],b.shape[1]),'d',order='F')
mylapack.dgemm('N','N',a.shape[0],b.shape[1],a.shape[1],1.0,a,b,1.0,c)
print(c)
np.all(c == a @ b) # check with numpy matrix multiplication
---------------------------------------------------------------------------
ImportError Traceback (most recent call last)
Cell In[24], line 2
1 import numpy as np
----> 2 import mylapack
3 a = np.array([[7,8],[3,4],[1,2]])
4 b = np.array([[1,2,3],[4,5,6]])
ImportError: /home/runner/work/python-fortran/python-fortran/notebooks/mylapack.cpython-39-x86_64-linux-gnu.so: undefined symbol: dgemm_
Exercise#
Modify the file dgemm.pyf to set all arguments top optional and keep only the two matrices as input.
%%file dgemm2.pyf
python module mylapack2 ! in
interface ! in :mylapack
subroutine dgemm(transa,transb,m,n,k,alpha,a,lda,b,ldb,beta,c,ldc)
character, optional :: transa = 'N'
character, optional :: transb = 'N'
integer, optional :: m = shape(a,0)
integer, optional :: n = shape(b,1)
integer, optional, check(shape(b,0)==k) :: k = shape(a,1)
double precision, optional :: alpha = 1.
double precision dimension(lda,*) :: a
integer optional,check(shape(a,0)==lda),depend(a) :: lda=shape(a,0)
double precision dimension(ldb,*) :: b
integer optional,check(shape(b,0)==ldb),depend(b) :: ldb=shape(b,0)
double precision, optional :: beta = 1.
double precision dimension(shape(a,0),shape(b,1)), intent(out) :: c
integer optional :: ldc=shape(a,0)
end subroutine dgemm
end interface
end python module mylapack2
Overwriting dgemm2.pyf
Build the python module#
!{sys.executable} -m numpy.f2py --quiet -c dgemm2.pyf -m mylapack2 -llapack --f90flags=-O3
/home/pnavaro/.julia/conda/3/x86_64/envs/f2py/lib/python3.11/site-packages/numpy/f2py/f2py2e.py:723: VisibleDeprecationWarning:
distutils has been deprecated since NumPy 1.26.x
Use the Meson backend instead, or generate wrappers without -c and use a custom build script
builder = build_backend(
WARN: Could not locate executable armflang
Removing build directory /tmp/tmpsv7r5y41
import mylapack2
a = np.array([[7,8],[3,4],[1,2]])
b = np.array([[1,2,3],[4,5,6]])
c = mylapack2.dgemm(a,b)
np.all( c == a @ b)
np.True_
Check performance between numpy and mylapack#
a = np.random.random((512,128))
b = np.random.random((128,512))
%timeit c = mylapack2.dgemm(a,b)
756 μs ± 2.17 μs per loop (mean ± std. dev. of 7 runs, 1,000 loops each)
%timeit c = a @ b
1.51 ms ± 210 μs per loop (mean ± std. dev. of 7 runs, 1,000 loops each)
Fortran arrays allocated in a subroutine share same memory in Python
%%fortran
module f90module
implicit none
real(8), dimension(:), allocatable :: farray
contains
subroutine init( n ) !Allocation du tableau farray
integer, intent(in) :: n
allocate(farray(n))
end subroutine init
end module f90module
f90module.init(10)
len(f90module.farray)
10
Numpy arrays allocated in Python passed to Fortran are already allocated
%%fortran
module f90module
implicit none
real(8), dimension(:), allocatable :: farray
contains
subroutine test_array( allocated_flag, array_size )
logical, intent(out) :: allocated_flag
integer, intent(out) :: array_size
allocated_flag = allocated(farray)
array_size = size(farray)
end subroutine test_array
end module f90module
f90module.farray = np.random.rand(10).astype(np.float64)
f90module.test_array()
(1, 10)
f2py + OpenMP#
import os, sys
%env OMP_NUM_THREADS=4
if sys.platform == "darwin":
os.environ["CC"] = "gcc-10"
os.environ["FC"] = "gfortran"
env: OMP_NUM_THREADS=4
%%fortran
subroutine hello( )
integer :: i
do i = 1, 4
call sleep(1)
end do
end subroutine
%%time
hello()
CPU times: user 24.1 ms, sys: 44.5 ms, total: 68.6 ms
Wall time: 4 s
%%fortran --f90flags='-fopenmp' --extra='-lgomp'
subroutine hello_omp( )
use omp_lib
integer :: i
!$OMP PARALLEL PRIVATE(I)
!$OMP DO
do i = 1, 4
call sleep(1)
end do
!$OMP END DO
!$OMP END PARALLEL
end subroutine hello_omp
%%time
hello_omp()
CPU times: user 8.49 ms, sys: 8.8 ms, total: 17.3 ms
Wall time: 1 s
Conclusions#
Easy to use, it works with modern fortran, legacy fortran and also C.
Works with common and modules and arrays dynamically allocated.
Python function callback can be very useful combined with Sympy
Documentation is automatically generated
All fortran compilers are supported: GNU, Portland, Sun, Intel,…
F2py is integrated in numpy library.
cons#
Derived types and fortran pointers are not well supported.
Absolutely not compatible with fortran 2003-2008 new features (classes)
f2py is maintained but not really improved. Development is stopped.
distutils#
from numpy.distutils.core import Extension, setup
ext1 = Extension(name = 'scalar',
sources = ['scalar.f'])
ext2 = Extension(name = 'fib2',
sources = ['fib2.pyf','fib1.f'])
setup(name = 'f2py_example', ext_modules = [ext1,ext2])
Compilation
python3 setup.py build_ext --inplace
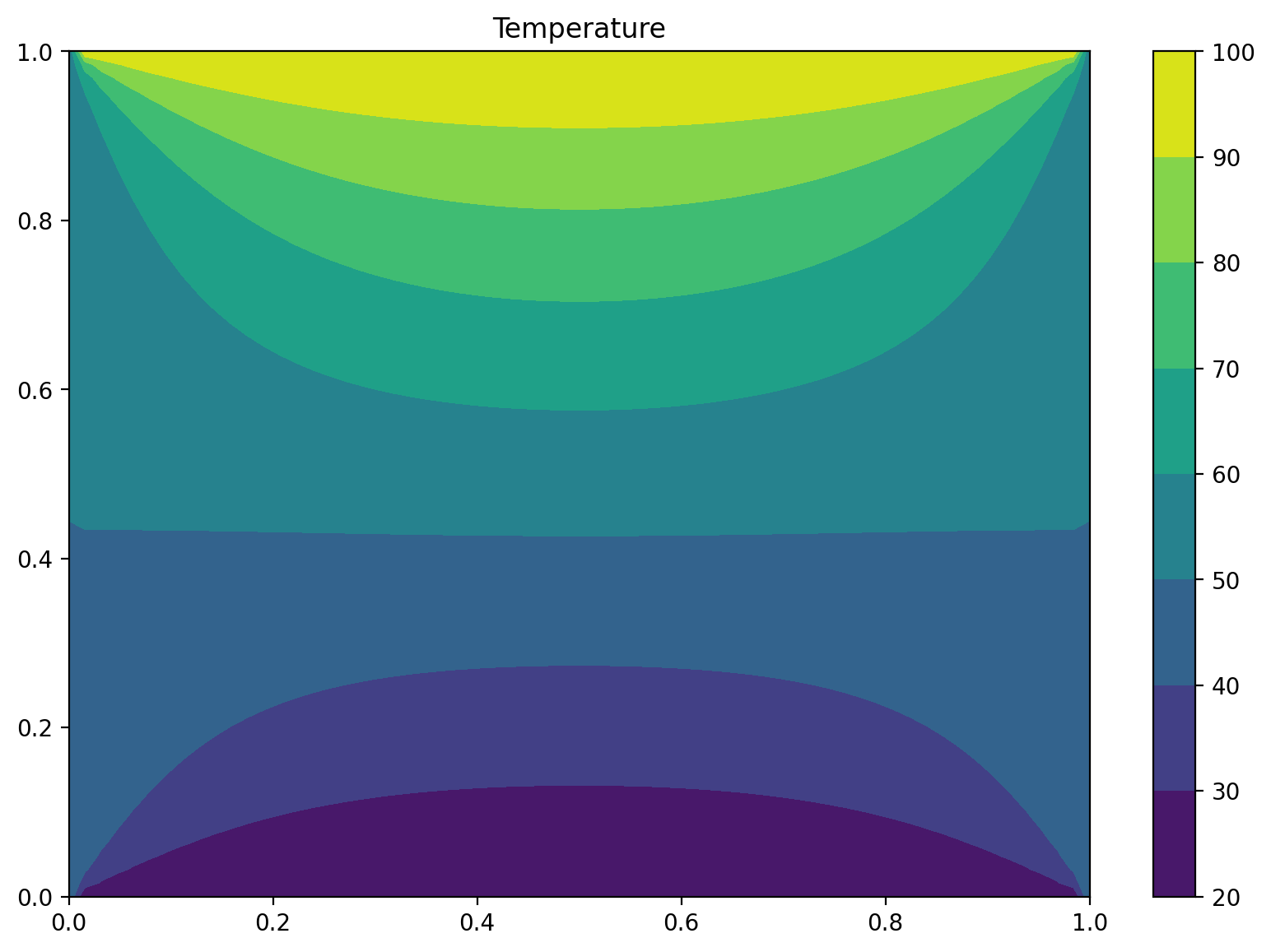
Exercice: Laplace problem#
Replace the
laplacefunction by a fortran subroutine
%%time
%config InlineBackend.figure_format = 'retina'
import numpy as np
import matplotlib.pyplot as plt
import itertools
# Boundary conditions
Tnorth, Tsouth, Twest, Teast = 100, 20, 50, 50
# Set meshgrid
n, l = 64, 1.0
X, Y = np.meshgrid(np.linspace(0,l,n), np.linspace(0,l,n))
T = np.zeros((n,n))
# Set Boundary condition
T[n-1:, :] = Tnorth
T[:1, :] = Tsouth
T[:, n-1:] = Teast
T[:, :1] = Twest
def laplace(T, n):
residual = 0.0
for i in range(1, n-1):
for j in range(1, n-1):
T_old = T[i,j]
T[i, j] = 0.25 * (T[i+1,j] + T[i-1,j] + T[i,j+1] + T[i,j-1])
if T[i,j]>0:
residual=max(residual,abs((T_old-T[i,j])/T[i,j]))
return residual
residual = 1.0
istep = 0
while residual > 1e-5 :
istep += 1
residual = laplace(T, n)
# print ((istep, residual), end="\r")
print("iterations = ",istep)
plt.rcParams['figure.figsize'] = (10,6.67)
plt.title("Temperature")
plt.contourf(X, Y, T)
plt.colorbar()
%%fortran
subroutine laplace_fortran( T, n, residual )
real(8), intent(inout) :: T(0:n-1,0:n-1) ! Python indexing
integer, intent(in) :: n
real(8), intent(out) :: residual
real(8) :: T_old
residual = 0.0
do i = 1, n-2
do j = 1, n-2
T_old = T(i,j)
T(i, j) = 0.25 * (T(i+1,j) + T(i-1,j) + T(i,j+1) + T(i,j-1))
if (T(i,j) > 0) then
residual=max(residual,abs((T_old-T(i,j))/T(i,j)))
end if
end do
end do
end subroutine laplace_fortran
%%time
%config InlineBackend.figure_format = 'retina'
import numpy as np
import matplotlib.pyplot as plt
import itertools
# Boundary conditions
Tnorth, Tsouth, Twest, Teast = 100, 20, 50, 50
# Set meshgrid
n, l = 64, 1.0
X, Y = np.meshgrid(np.linspace(0,l,n), np.linspace(0,l,n))
T = np.zeros((n,n), order='F') ## We need to declare a new order in memory
# Set Boundary condition
T[n-1:, :] = Tnorth
T[:1, :] = Tsouth
T[:, n-1:] = Teast
T[:, :1] = Twest
residual = 1.0
istep = 0
while residual > 1e-5 :
istep += 1
residual = laplace_fortran(T, n)
print()
print("iterations = ",istep)
plt.rcParams['figure.figsize'] = (10,6.67)
plt.title("Temperature")
plt.contourf(X, Y, T)
plt.colorbar()
References#
Hans Petter Langtangen. Python Scripting for Computational Science. Springer 2004
Add extra flags like bounds checking#
python3 setup.py config_fc --f90flags=-fbounds-check
In setup.py
ext1 = Extension(name='_coffee_f90',
sources=files1,
extra_f90_compile_args=['-fbounds-check'],
extra_link_args=['-llapack -lgomp'])