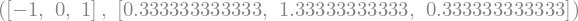Sympy#
%matplotlib inline
%config InlineBackend.figure_format = "retina"
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
sns.set()
plt.rcParams['figure.figsize'] = (10.0, 6.0)

The function init_printing() will enable LaTeX pretty printing in the notebook for SymPy expressions.
import sympy as sym
from sympy import symbols, Symbol
sym.init_printing()
x= Symbol('x')
(sym.pi + x)**2
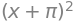
alpha1, omega_2 = symbols('alpha1 omega_2')
alpha1, omega_2

mu, sigma = sym.symbols('mu sigma', positive = True)
1/sym.sqrt(2*sym.pi*sigma**2)* sym.exp(-(x-mu)**2/(2*sigma**2))
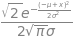
Why use sympy?#
Symbolic derivatives
Translate mathematics into low level code
Deal with very large expressions
Optimize code using mathematics
Dividing two integers in Python creates a float, like 1/2 -> 0.5. If you want a rational number, use Rational(1, 2) or S(1)/2.
x + sym.S(1)/2 , sym.Rational(1,4)

y = Symbol('y')
x ^ y # XOR operator (True only if x != y)
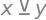
x**y

SymPy expressions are immutable. Functions that operate on an expression return a new expression.
expr = x + 1
expr

expr.subs(x, 2)
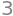
expr

Exercise: Lagrange polynomial#
Given a set of \(k + 1\) data points :\((x_0, y_0),\ldots,(x_j, y_j),\ldots,(x_k, y_k)\) the Lagrange interpolation polynomial is:
\(\ell_j\) are Lagrange basis polynomials: $\(\ell_j(x) := \prod_{\begin{smallmatrix}0\le m\le k\\ m\neq j\end{smallmatrix}} \frac{x-x_m}{x_j-x_m} \)\( We can demonstrate that at each point \)x_i\(, \)L(x_i)=y_i\( so \)L$ interpolates the function.
Compute the Lagrange polynomial for points $\( (-2,21),(-1,1),(0,-1),(1,-3),(2,1) \)$
Evaluate an expression#
sym.sqrt(2), sym.sqrt(2).evalf(7) # set the precision to 7 digits
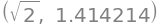
from sympy import sin
x = Symbol('x')
expr = sin(x)/x
expr.evalf(subs={x: 3.14}) # substitute the symbol x by Pi value
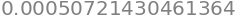
from sympy.utilities.autowrap import ufuncify
f = ufuncify([x], expr, backend='f2py')
t = np.linspace(0,4*np.pi,100)
plt.plot(t, f(t));
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
File ~/miniconda3/envs/runenv/lib/python3.13/site-packages/sympy/core/cache.py:72, in __cacheit.<locals>.func_wrapper.<locals>.wrapper(*args, **kwargs)
71 try:
---> 72 retval = cfunc(*args, **kwargs)
73 except TypeError as e:
TypeError: unhashable type: 'list'
During handling of the above exception, another exception occurred:
TypeError Traceback (most recent call last)
File ~/miniconda3/envs/runenv/lib/python3.13/site-packages/sympy/core/cache.py:72, in __cacheit.<locals>.func_wrapper.<locals>.wrapper(*args, **kwargs)
71 try:
---> 72 retval = cfunc(*args, **kwargs)
73 except TypeError as e:
TypeError: unhashable type: 'list'
During handling of the above exception, another exception occurred:
CalledProcessError Traceback (most recent call last)
File ~/miniconda3/envs/runenv/lib/python3.13/site-packages/sympy/utilities/autowrap.py:183, in CodeWrapper._process_files(self, routine)
182 try:
--> 183 retoutput = check_output(command, stderr=STDOUT)
184 except CalledProcessError as e:
File ~/miniconda3/envs/runenv/lib/python3.13/subprocess.py:472, in check_output(timeout, *popenargs, **kwargs)
470 kwargs['input'] = empty
--> 472 return run(*popenargs, stdout=PIPE, timeout=timeout, check=True,
473 **kwargs).stdout
File ~/miniconda3/envs/runenv/lib/python3.13/subprocess.py:577, in run(input, capture_output, timeout, check, *popenargs, **kwargs)
576 if check and retcode:
--> 577 raise CalledProcessError(retcode, process.args,
578 output=stdout, stderr=stderr)
579 return CompletedProcess(process.args, retcode, stdout, stderr)
CalledProcessError: Command '['/home/runner/miniconda3/envs/runenv/bin/python', '-c', 'import numpy.f2py as f2py2e;f2py2e.main()', '-c', '-m', 'wrapper_module_0', 'wrapped_code_0.f90']' returned non-zero exit status 1.
During handling of the above exception, another exception occurred:
CodeWrapError Traceback (most recent call last)
Cell In[14], line 2
1 from sympy.utilities.autowrap import ufuncify
----> 2 f = ufuncify([x], expr, backend='f2py')
4 t = np.linspace(0,4*np.pi,100)
5 plt.plot(t, f(t));
File ~/miniconda3/envs/runenv/lib/python3.13/site-packages/sympy/core/cache.py:76, in __cacheit.<locals>.func_wrapper.<locals>.wrapper(*args, **kwargs)
74 if not e.args or not e.args[0].startswith('unhashable type:'):
75 raise
---> 76 retval = func(*args, **kwargs)
77 return retval
File ~/miniconda3/envs/runenv/lib/python3.13/site-packages/sympy/utilities/autowrap.py:1177, in ufuncify(args, expr, language, backend, tempdir, flags, verbose, helpers, **kwargs)
1175 args = [y] + indexed_args + [m]
1176 args_with_indices = [a[i] for a in indexed_args]
-> 1177 return autowrap(Eq(y[i], f(*args_with_indices)), language, backend,
1178 tempdir, args, flags, verbose, helpers, **kwargs)
File ~/miniconda3/envs/runenv/lib/python3.13/site-packages/sympy/core/cache.py:76, in __cacheit.<locals>.func_wrapper.<locals>.wrapper(*args, **kwargs)
74 if not e.args or not e.args[0].startswith('unhashable type:'):
75 raise
---> 76 retval = func(*args, **kwargs)
77 return retval
File ~/miniconda3/envs/runenv/lib/python3.13/site-packages/sympy/utilities/autowrap.py:690, in autowrap(expr, language, backend, tempdir, args, flags, verbose, helpers, code_gen, **kwargs)
687 new_args.append(missing.name)
688 routine = code_gen.routine('autofunc', expr, args + new_args)
--> 690 return code_wrapper.wrap_code(routine, helpers=helps)
File ~/miniconda3/envs/runenv/lib/python3.13/site-packages/sympy/utilities/autowrap.py:164, in CodeWrapper.wrap_code(self, routine, helpers)
162 self._generate_code(routine, helpers)
163 self._prepare_files(routine)
--> 164 self._process_files(routine)
165 mod = __import__(self.module_name)
166 finally:
File ~/miniconda3/envs/runenv/lib/python3.13/site-packages/sympy/utilities/autowrap.py:185, in CodeWrapper._process_files(self, routine)
183 retoutput = check_output(command, stderr=STDOUT)
184 except CalledProcessError as e:
--> 185 raise CodeWrapError(
186 "Error while executing command: %s. Command output is:\n%s" % (
187 " ".join(command), e.output.decode('utf-8')))
188 if not self.quiet:
189 print(retoutput)
CodeWrapError: Error while executing command: /home/runner/miniconda3/envs/runenv/bin/python -c import numpy.f2py as f2py2e;f2py2e.main() -c -m wrapper_module_0 wrapped_code_0.f90. Command output is:
Traceback (most recent call last):
File "<string>", line 1, in <module>
import numpy.f2py as f2py2e;f2py2e.main()
~~~~~~~~~~~^^
File "/home/runner/miniconda3/envs/runenv/lib/python3.13/site-packages/numpy/f2py/f2py2e.py", line 781, in main
run_compile()
~~~~~~~~~~~^^
File "/home/runner/miniconda3/envs/runenv/lib/python3.13/site-packages/numpy/f2py/f2py2e.py", line 753, in run_compile
builder.compile()
~~~~~~~~~~~~~~~^^
File "/home/runner/miniconda3/envs/runenv/lib/python3.13/site-packages/numpy/f2py/_backends/_meson.py", line 192, in compile
self.run_meson(self.build_dir)
~~~~~~~~~~~~~~^^^^^^^^^^^^^^^^
File "/home/runner/miniconda3/envs/runenv/lib/python3.13/site-packages/numpy/f2py/_backends/_meson.py", line 185, in run_meson
self._run_subprocess_command(setup_command, build_dir)
~~~~~~~~~~~~~~~~~~~~~~~~~~~~^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/runner/miniconda3/envs/runenv/lib/python3.13/site-packages/numpy/f2py/_backends/_meson.py", line 181, in _run_subprocess_command
subprocess.run(command, cwd=cwd, check=True)
~~~~~~~~~~~~~~^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/runner/miniconda3/envs/runenv/lib/python3.13/subprocess.py", line 554, in run
with Popen(*popenargs, **kwargs) as process:
~~~~~^^^^^^^^^^^^^^^^^^^^^^
File "/home/runner/miniconda3/envs/runenv/lib/python3.13/subprocess.py", line 1039, in __init__
self._execute_child(args, executable, preexec_fn, close_fds,
~~~~~~~~~~~~~~~~~~~^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
pass_fds, cwd, env,
^^^^^^^^^^^^^^^^^^^
...<5 lines>...
gid, gids, uid, umask,
^^^^^^^^^^^^^^^^^^^^^^
start_new_session, process_group)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/runner/miniconda3/envs/runenv/lib/python3.13/subprocess.py", line 1972, in _execute_child
raise child_exception_type(errno_num, err_msg, err_filename)
FileNotFoundError: [Errno 2] No such file or directory: 'meson'
Cannot use distutils backend with Python>=3.12, using meson backend instead.
Using meson backend
Will pass --lower to f2py
See https://numpy.org/doc/stable/f2py/buildtools/meson.html
Reading fortran codes...
Reading file 'wrapped_code_0.f90' (format:free)
Post-processing...
Block: wrapper_module_0
Block: autofunc
Applying post-processing hooks...
character_backward_compatibility_hook
Post-processing (stage 2)...
Building modules...
Building module "wrapper_module_0"...
Generating possibly empty wrappers"
Maybe empty "wrapper_module_0-f2pywrappers.f"
Constructing wrapper function "autofunc"...
y_5262565 = autofunc(x_5262569,[m_5262566])
Wrote C/API module "wrapper_module_0" to file "./wrapper_module_0module.c"
Exercise#
Plot the Lagrange polynomial computed above and interpolations points with matplotlib
Undefined functions and derivatives#
Undefined functions are created with Function(). Undefined are useful to state that one variable depends on another (for the purposes of differentiation).
from sympy import Function
f = Function('f')
f(x) + 1
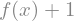
from sympy import diff, sin, cos
diff(sin(x + 1)*cos(y), x), diff(sin(x + 1)*cos(y), x, y), diff(f(x), x)

c, t = sym.symbols('t c')
u = sym.Function('u')
sym.Eq(diff(u(t,x),t,t), c**2*diff(u(t,x),x,2))
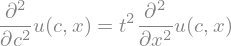
Matrices#
from sympy import Matrix
Matrix([[1, 2], [3, 4]])*Matrix([x, y])
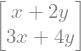
x, y, z = sym.symbols('x y z')
Matrix([sin(x) + y, cos(y) + x, z]).jacobian([x, y, z])
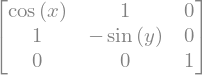
Matrix symbols#
SymPy can also operate on matrices of symbolic dimension (\(n \times m\)). MatrixSymbol("M", n, m) creates a matrix \(M\) of shape \(n \times m\).
from sympy import MatrixSymbol, Transpose
n, m = sym.symbols('n m', integer=True)
M = MatrixSymbol("M", n, m)
b = MatrixSymbol("b", m, 1)
Transpose(M*b)
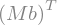
Transpose(M*b).doit()
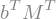
Solving systems of equations#
solve solves equations symbolically (not numerically). The return value is a list of solutions. It automatically assumes that it is equal to 0.
from sympy import Eq, solve
solve(Eq(x**2, 4), x)
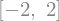
solve(x**2 + 3*x - 3, x)
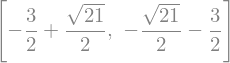
eq1 = x**2 + y**2 - 4 # circle of radius 2
eq2 = 2*x + y - 1 # straight line: y(x) = -2*x + 1
solve([eq1, eq2], [x, y])
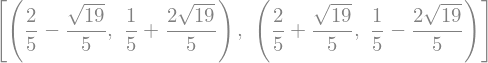
Solving differential equations#
dsolve can (sometimes) produce an exact symbolic solution. Like solve, dsolve assumes that expressions are equal to 0.
from sympy import Function, dsolve
f = Function('f')
dsolve(f(x).diff(x, 2) + f(x))
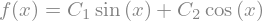
Code printers#
The most basic form of code generation are the code printers. They convert SymPy expressions into over a dozen target languages.
x = symbols('x')
expr = abs(sin(x**2))
expr
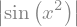
sym.ccode(expr)
'fabs(sin(pow(x, 2)))'
sym.fcode(expr, standard=2003, source_format='free')
'abs(sin(x**2))'
from sympy.printing.cxxcode import cxxcode
cxxcode(expr)
'std::fabs(std::sin(std::pow(x, 2)))'
sym.tanh(x).rewrite(sym.exp)
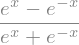
from sympy import sqrt, exp, pi
expr = 1/sqrt(2*pi*sigma**2)* exp(-(x-mu)**2/(2*sigma**2))
print(sym.fcode(expr, standard=2003, source_format='free'))
parameter (pi = 3.1415926535897932d0)
(1.0d0/2.0d0)*sqrt(2.0d0)*exp(-0.5d0*(-mu + x)**2/sigma**2)/(sqrt(pi)* &
sigma)
Creating a function from a symbolic expression#
In SymPy there is a function to create a Python function which evaluates (usually numerically) an expression. SymPy allows the user to define the signature of this function (which is convenient when working with e.g. a numerical solver in scipy).
from sympy import log
x, y = symbols('x y')
expr = 3*x**2 + log(x**2 + y**2 + 1)
expr
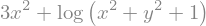
%timeit expr.subs({x: 17, y: 42}).evalf()
478 µs ± 50.7 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
import math
f = lambda x, y: 3*x**2 + math.log(x**2 + y**2 + 1)
f(17, 42)
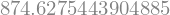
%timeit f(17, 42)
3.84 µs ± 614 ns per loop (mean ± std. dev. of 7 runs, 100000 loops each)
Evaluate above expression numerically invoking the subs method followed by the evalf method can be quite slow and cannot be done repeatedly.
from sympy import lambdify
g = lambdify([x, y], expr, modules=['math'])
g(17, 42)
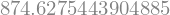
%timeit g(17, 42)
3.67 µs ± 715 ns per loop (mean ± std. dev. of 7 runs, 100000 loops each)
xarr = np.linspace(17, 18, 5)
h = lambdify([x, y], expr) # lambdify return a python function
out = h(xarr, 42)
out.shape

z = z1, z2, z3 = symbols('z:3')
expr2 = x*y*(z1 + z2 + z3)
func2 = lambdify([x, y, z], expr2)
func2(1, 2, (3, 4, 5))
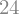
Behind the scenes lambdify constructs a string representation of the Python code and uses Python’s eval function to compile the function.
SIR model#
\begin{align} \frac{dS(t)}{dt} &= - \beta S(t) I(t) \ \frac{dI(t)}{dt} &= \beta S(t) I(t) - \gamma I(t) \ \frac{dR(t)}{dt} &= \gamma I(t) \end{align}
S,I,R: ratio of suceptibles, infectious and recovered fraction of the population.
t: time
\(\beta\) : transmission coefficient.
\(\gamma\) : healing rate.
We assume that total population is constant.
Solving the initial value problem numerically#
We will now integrate this system of ordinary differential equations numerically using the odeint solver provided by scipy:
By looking at the documentation of odeint we see that we need to provide a function which computes a vector of derivatives (\(\dot{\mathbf{y}} = [\frac{dy_1}{dt}, \frac{dy_2}{dt}, \frac{dy_3}{dt}]\)). The expected signature of this function is:
f(y: array[float64], t: float64, *args: arbitrary constants) -> dydt: array[float64]
in our case we can write it as:
def rhs(y, t, beta, gamma):
rb = beta * y[0]*y[1]
rg = gamma * y[1]
return [- rb , rb - rg, rg]
import scipy.integrate as spi
tout = np.linspace(0, 10, 100)
k_vals = 1.66, 0.4545455
y0 = [0.95, 0.05, 0]
yout = spi.odeint(rhs, y0, tout, k_vals)
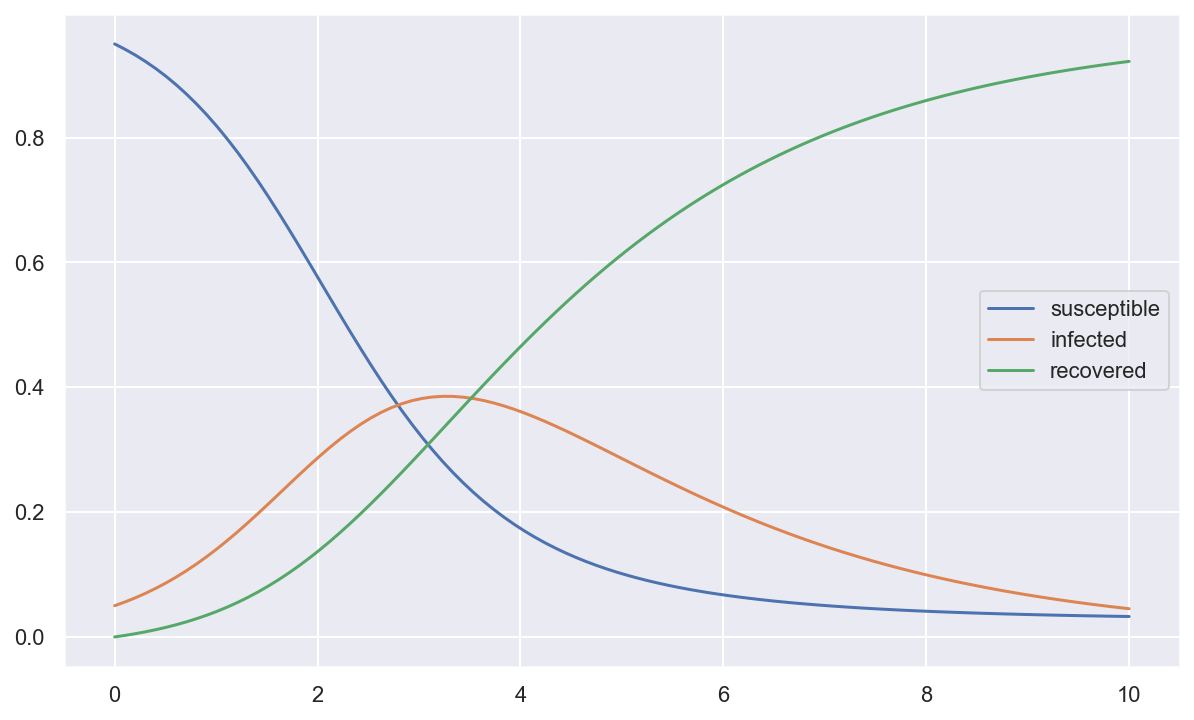
plt.plot(tout, yout)
plt.legend(['susceptible', 'infected', 'recovered']);
We will construct the system from a symbolic representation. But at the same time, we need the rhs function to be fast. Which means that we want to produce a fast function from our symbolic representation. Generating a function from our symbolic representation is achieved through code generation.
Construct a symbolic representation from some domain specific representation using SymPy.
Have SymPy generate a function with an appropriate signature (or multiple thereof), which we pass on to the solver.
We will achieve (1) by using SymPy symbols (and functions if needed). For (2) we will use a function in SymPy called lambdify―it takes a symbolic expressions and returns a function. In a later notebook, we will look at (1), for now we will just use rhs which we’ve already written:
y, k = sym.symbols('y:3'), sym.symbols('beta gamma')
ydot = rhs(y, None, *k)
y, ydot
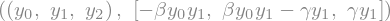
f = sym.lambdify((y,t)+k, ydot)
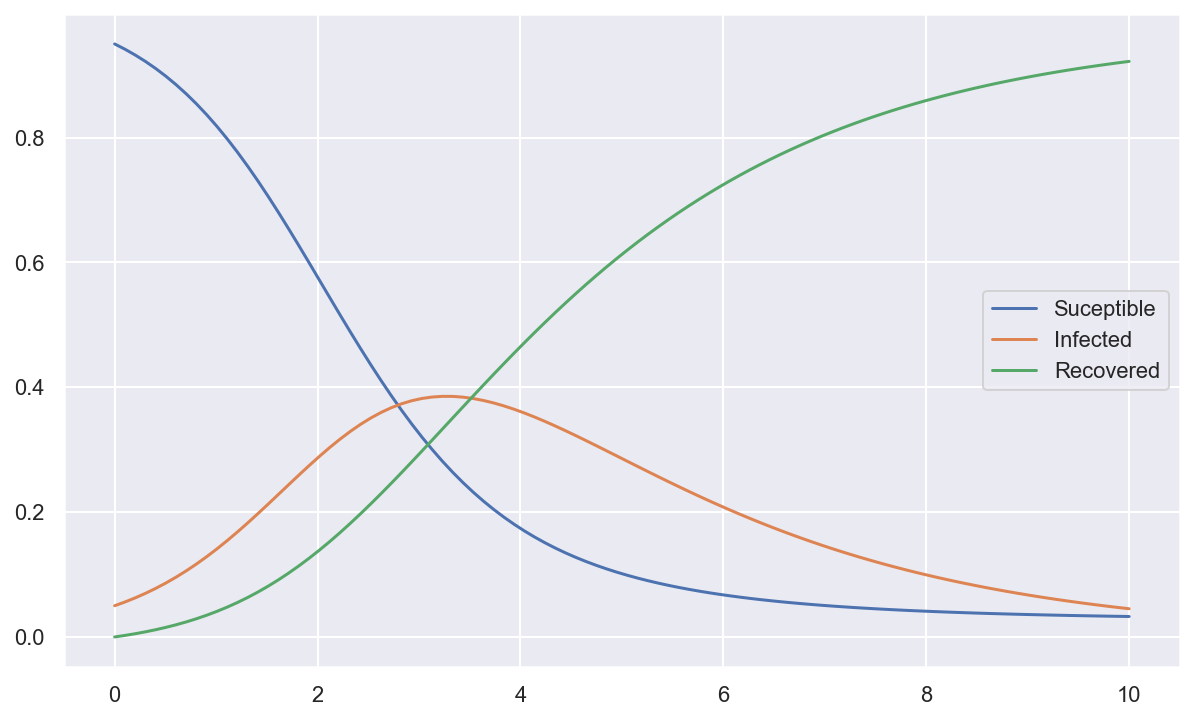
plt.plot(tout, spi.odeint(f, y0, tout, k_vals))
plt.legend(['Suceptible', 'Infected', 'Recovered']);
In this example the gains of using a symbolic representation are arguably limited.
Let’s take the same example with demography and \(n\) classes of subjects:
\(\beta\) : transmission coefficient
\(\gamma\) : healing rate
\(\mu\) : mortality rate
\(\nu\) : birth rate
Exercise#
Create the symbolic matrix \(m\), symbols \(\nu_i,\mu_i,\beta_i,\gamma_i\) for \(i=0,1,2\) and \(y_j\) for \(j=0,1,2,\ldots,8\)
Write the system \(\dot{y} = f(t,y,m,\nu,\mu,\beta,\gamma)\)
lambdifythe \(f\) function.Solve the system with: $\( m = \begin{bmatrix} 0 & 0.01 & 0.01 \\ 0.01 & 0 & 0.01 \\ 0.01 & 0.01 & 0 \\ \end{bmatrix} \)\( \)\( \begin{aligned} t &= [0,10] \mbox{ with } dt = 0.1 \\ \nu_i &= 0.0 \\ \mu_i &= 0.0 \\ \beta_i &= 1.66 \\ \gamma_i &= [0.4545,0.3545,0.2545] \\ S_i&= 0.95 \\ I_i &= 0.05 \\ R_i &= 0.0 \end{aligned} \)$
Exercise : Bezier curve#
We want to compute and the draw the Bezier curve between the 3 points \(p_0\), \(p_1\), and \(p_2\), The middle point \(p_1\) position is arbitrary.
The \(n+1\) Bernstein basis polynomials of degree \(n\) are defined as
where \({n \choose i}\) is the binomial coefficient.
The Bezier curve is defined by a linear combination of Bernstein basis polynomials:
With
sympy.binomial, write a functionbpoly(t,n,i)that returns the Bernstein basis polynomial \(b_{i,n}(t)\).Compute the Berstein polynomial representing the Bezier curve between \(p_0,p_1,p_2\). \(\beta_i=1\).
Plot the Bezier Curve for 3 positions of \(p_1= (0,0), (0.5,0.5), (1,1)\)
Integrals quadrature#
from sympy.integrals.quadrature import *
x, w = gauss_legendre(3, 5)
x, w
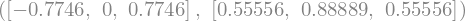
x, w = gauss_lobatto(3,12)
x, w