Scipy#

Scipy is the scientific Python ecosystem :
fft, linear algebra, scientific computation,…
scipy contains numpy, it can be considered as an extension of numpy.
the add-on toolkits Scikits complements scipy.
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import numpy as np
import scipy as sp
import matplotlib.pyplot as plt
plt.rcParams['figure.figsize'] = (10,6)
SciPy main packages#
constants: Physical and mathematical constantsfftpack: Fast Fourier Transform routinesintegrate: Integration and ordinary differential equation solversinterpolate: Interpolation and smoothing splinesio: Input and Outputlinalg: Linear algebrasignal: Signal processingsparse: Sparse matrices and associated routines
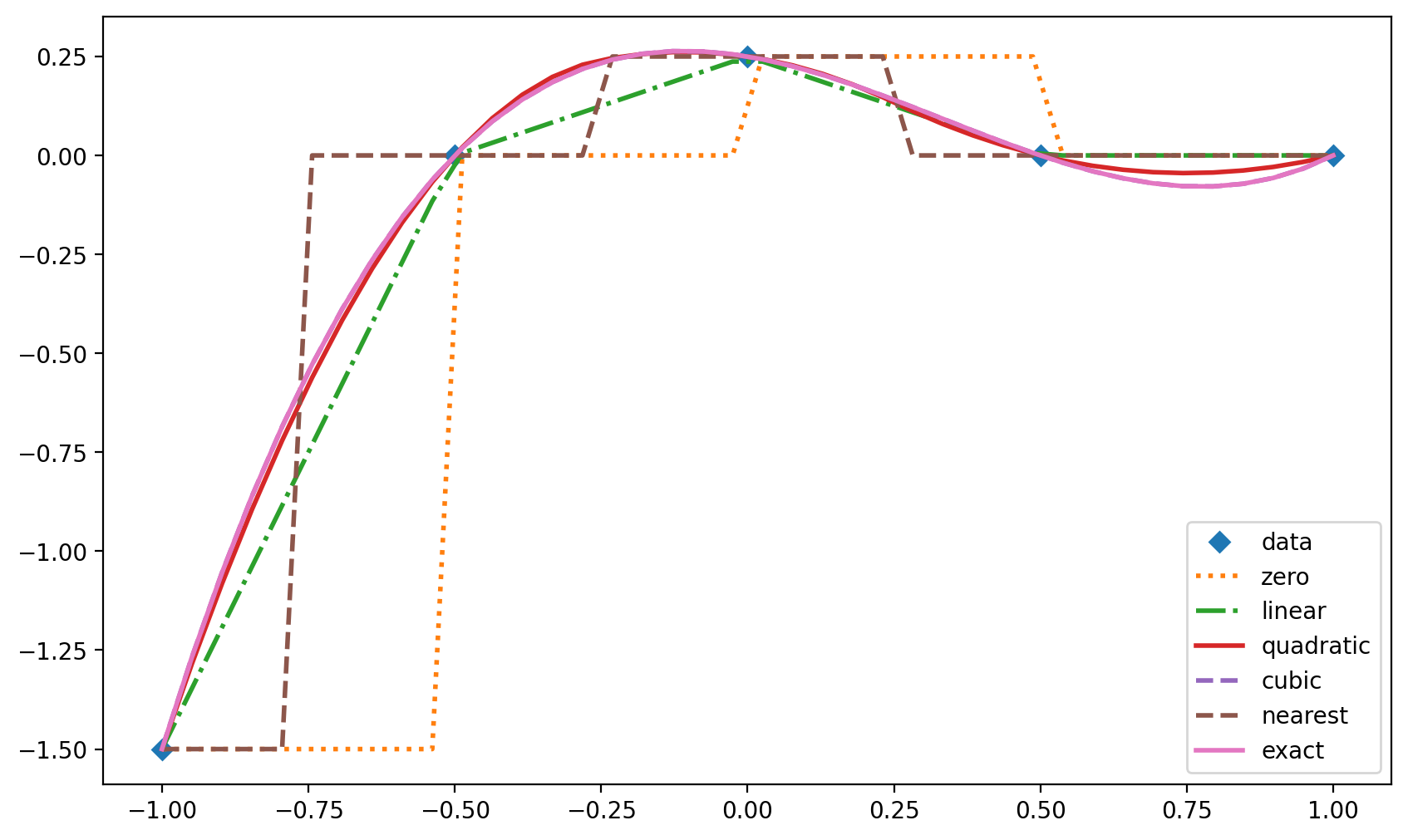
from scipy.interpolate import interp1d
x = np.linspace(-1, 1, num=5) # 5 points evenly spaced in [-1,1].
y = (x-1.)*(x-0.5)*(x+0.5) # x and y are numpy arrays
f0 = interp1d(x,y, kind='zero')
f1 = interp1d(x,y, kind='linear')
f2 = interp1d(x,y, kind='quadratic')
f3 = interp1d(x,y, kind='cubic')
f4 = interp1d(x,y, kind='nearest')
xnew = np.linspace(-1, 1, num=40)
ynew = (xnew-1.)*(xnew-0.5)*(xnew+0.5)
plt.plot(x,y,'D',xnew,f0(xnew),':', xnew, f1(xnew),'-.',
xnew,f2(xnew),'-',xnew ,f3(xnew),'--',
xnew,f4(xnew),'--',xnew, ynew, linewidth=2)
plt.legend(['data','zero','linear','quadratic','cubic','nearest','exact'],
loc='best');
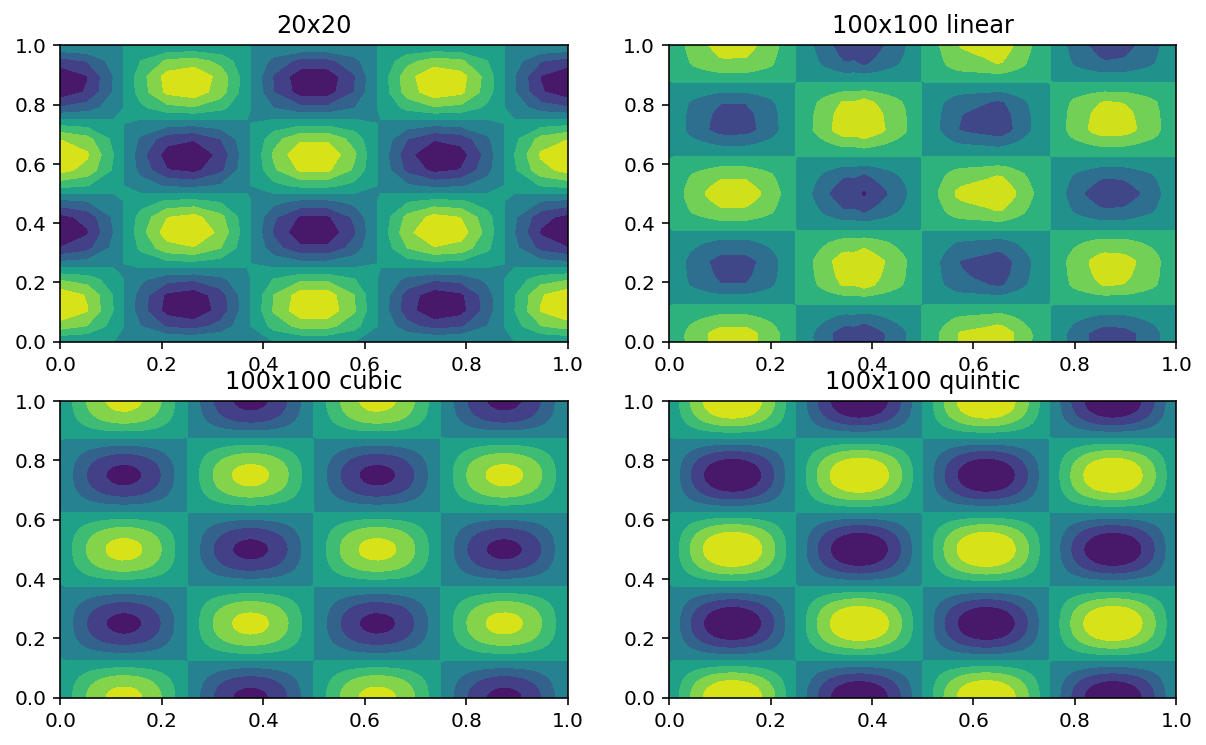
from scipy.interpolate import interp2d
x,y = sp.mgrid[0:1:20j,0:1:20j] #create the grid 20x20
z = np.cos(4*sp.pi*x)*np.sin(4*sp.pi*y) #initialize the field
T1=interp2d(x,y,z,kind='linear')
T2=interp2d(x,y,z,kind='cubic')
T3=interp2d(x,y,z,kind='quintic')
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
File ~/miniconda3/envs/runenv/lib/python3.13/site-packages/scipy/__init__.py:134, in __getattr__(name)
133 try:
--> 134 return globals()[name]
135 except KeyError:
KeyError: 'mgrid'
During handling of the above exception, another exception occurred:
AttributeError Traceback (most recent call last)
Cell In[4], line 2
1 from scipy.interpolate import interp2d
----> 2 x,y = sp.mgrid[0:1:20j,0:1:20j] #create the grid 20x20
3 z = np.cos(4*sp.pi*x)*np.sin(4*sp.pi*y) #initialize the field
4 T1=interp2d(x,y,z,kind='linear')
File ~/miniconda3/envs/runenv/lib/python3.13/site-packages/scipy/__init__.py:136, in __getattr__(name)
134 return globals()[name]
135 except KeyError:
--> 136 raise AttributeError(
137 f"Module 'scipy' has no attribute '{name}'"
138 )
AttributeError: Module 'scipy' has no attribute 'mgrid'
X,Y=sp.mgrid[0:1:100j,0:1:100j] #create the interpolation grid 100x100
# complex -> number of points, float -> step size
plt.figure(1)
plt.subplot(221) #Plot original data
plt.contourf(x,y,z)
plt.title('20x20')
plt.subplot(222) #Plot linear interpolation
plt.contourf(X,Y,T1(X[:,0],Y[0,:]))
plt.title('100x100 linear')
plt.subplot(223) #Plot cubic interpolation
plt.contourf(X,Y,T2(X[:,0],Y[0,:]))
plt.title('100x100 cubic')
plt.subplot(224) #Plot quintic interpolation
plt.contourf(X,Y,T3(X[:,0],Y[0,:]))
plt.title('100x100 quintic')
FFT : scipy.fftpack#
FFT dimension 1, 2 and n : fft, ifft (inverse), rfft (real), irfft, fft2 (dimension 2), ifft2, fftn (dimension n), ifftn.
Discrete cosinus transform : dct
Convolution product : convolve
from numpy.fft import fft, ifft
x = np.random.random(2048)
%timeit ifft(fft(x))
102 µs ± 2.08 µs per loop (mean ± std. dev. of 7 runs, 10000 loops each)
from scipy.fftpack import fft, ifft
x = np.random.random(2048)
%timeit ifft(fft(x))
90 µs ± 567 ns per loop (mean ± std. dev. of 7 runs, 10000 loops each)
Linear algebra : scipy.linalg#
Sovers, decompositions, eigen values. (same as numpy).
Matrix functions : expm, sinm, sinhm,…
Block matrices diagonal, triangular, periodic,…
import scipy.linalg as spl
b=np.ones(5)
A=np.array([[1.,3.,0., 0.,0.],
[ 2.,1.,-4, 0.,0.],
[ 6.,1., 2,-3.,0.],
[ 0.,1., 4.,-2.,-3.],
[ 0.,0., 6.,-3., 2.]])
print("x=",spl.solve(A,b,sym_pos=False)) # LAPACK ( gesv ou posv )
AB=np.array([[0.,3.,-4.,-3.,-3.],
[1.,1., 2.,-2., 2.],
[2.,1., 4.,-3., 0.],
[6.,1., 6., 0., 0.]])
print("x=",spl.solve_banded((2,1),AB,b)) # LAPACK ( gbsv )
x= [-0.24074074 0.41358025 -0.26697531 -0.85493827 0.01851852]
x= [-0.24074074 0.41358025 -0.26697531 -0.85493827 0.01851852]
P,L,U = spl.lu(A) # P A = L U
np.set_printoptions(precision=3)
for M in (P,L,U):
print(M, end="\n"+20*"-"+"\n")
[[0. 1. 0. 0. 0.]
[0. 0. 0. 1. 0.]
[1. 0. 0. 0. 0.]
[0. 0. 0. 0. 1.]
[0. 0. 1. 0. 0.]]
--------------------
[[ 1. 0. 0. 0. 0. ]
[ 0.167 1. 0. 0. 0. ]
[ 0. 0. 1. 0. 0. ]
[ 0.333 0.235 -0.765 1. 0. ]
[ 0. 0.353 0.686 0.083 1. ]]
--------------------
[[ 6. 1. 2. -3. 0. ]
[ 0. 2.833 -0.333 0.5 0. ]
[ 0. 0. 6. -3. 2. ]
[ 0. 0. 0. -1.412 1.529]
[ 0. 0. 0. 0. -4.5 ]]
--------------------
CSC (Compressed Sparse Column)#
All operations are optimized
Efficient “slicing” along axis=1.
Fast Matrix-vector product.
Conversion to other format could be costly.
import scipy.sparse as spsp
row = np.array([0,2,2,0,1,2])
col = np.array([0,0,1,2,2,2])
data = np.array([1,2,3,4,5,6])
Mcsc1 = spsp.csc_matrix((data,(row,col)),shape=(3,3))
Mcsc1.todense()
matrix([[1, 0, 4],
[0, 0, 5],
[2, 3, 6]])
indptr = np.array([0,2,3,6])
indices = np.array([0,2,2,0,1,2])
data = np.array([1,2,3,4,5,6])
Mcsc2 = spsp.csc_matrix ((data,indices,indptr),shape=(3,3))
Mcsc2.todense()
matrix([[1, 0, 4],
[0, 0, 5],
[2, 3, 6]])
Dedicated format for assembling#
lil_matrix: Row-based linked list matrix. Easy format to build your matrix and convert to other format before solving.dok_matrix: A dictionary of keys based matrix. Ideal format for incremental matrix building. The conversion to csc/csr format is efficient.coo_matrix: coordinate list format. Fast conversion to formats CSC/CSR.
Sparse matrices : scipy.sparse.linalg#
speigen, speigen_symmetric, lobpcg : (ARPACK).
svd : (ARPACK).
Direct methods (UMFPACK or SUPERLU) or Krylov based methods
Minimization : lsqr and minres
For linear algebra:
Noobs: spsolve.
Intermmediate: dsolve.spsolve or isolve.spsolve
Advanced: splu, spilu (direct); cg, cgs, bicg, bicgstab, gmres, lgmres et qmr (iterative)
Boss: petsc4py et slepc4py.
LinearOperator#
The LinearOperator is used for matrix-free numerical methods.
import scipy.sparse.linalg as spspl
def mv(v):
return np.array([2*v[0],3*v[1]])
A=spspl.LinearOperator((2 ,2),matvec=mv,dtype=float )
A
<2x2 _CustomLinearOperator with dtype=float64>
A*np.ones(2)
array([2., 3.])
A.matmat(np.array([[1,-2],[3,6]]))
array([[ 2, -4],
[ 9, 18]])
LU decomposition#
N = 50
un = np.ones(N)
w = np.random.rand(N+1)
A = spsp.spdiags([w[1:],-2*un,w[:-1]],[-1,0,1],N,N) # tridiagonal matrix
A = A.tocsc()
b = un
op = spspl.splu(A)
op
<SuperLU at 0x11d8bd1e0>
x=op.solve(b)
spl.norm(A*x-b)
2.3208707134311084e-15
Conjugate Gradient#
global k
k=0
def f(xk): # function called at every iterations
global k
print ("iteration {0:2d} residu = {1:7.3g}".format(k,spl.norm(A*xk-b)))
k += 1
x,info=spspl.cg(A,b,x0=np.zeros(N),tol=1.0e-12,maxiter=N,M=None,callback=f)
iteration 0 residu = 3.29
iteration 1 residu = 1.81
iteration 2 residu = 0.801
iteration 3 residu = 0.288
iteration 4 residu = 0.155
iteration 5 residu = 0.0755
iteration 6 residu = 0.0329
iteration 7 residu = 0.0126
iteration 8 residu = 0.00509
iteration 9 residu = 0.00211
iteration 10 residu = 0.00121
iteration 11 residu = 0.000495
iteration 12 residu = 0.00028
iteration 13 residu = 0.000105
iteration 14 residu = 4.49e-05
iteration 15 residu = 1.23e-05
iteration 16 residu = 5.31e-06
iteration 17 residu = 1.86e-06
iteration 18 residu = 9.85e-07
iteration 19 residu = 3.9e-07
iteration 20 residu = 1.78e-07
iteration 21 residu = 7.37e-08
iteration 22 residu = 2.27e-08
iteration 23 residu = 8.5e-09
iteration 24 residu = 2.04e-09
iteration 25 residu = 5.94e-10
iteration 26 residu = 1.11e-10
iteration 27 residu = 4.25e-11
iteration 28 residu = 7.55e-12
iteration 29 residu = 1.48e-12
Preconditioned conjugate gradient#
pc=spspl.spilu(A,drop_tol=0.1) # pc is an ILU decomposition
xp=pc.solve(b)
spl.norm(A*xp-b)
0.40994379196249386
def mv(v):
return pc.solve(v)
lo = spspl.LinearOperator((N,N),matvec=mv)
k = 0
x,info=spspl.cg(A,b,x0=np.zeros(N),tol=1.e-12,maxiter=N,M=lo,callback=f)
iteration 0 residu = 0.392
iteration 1 residu = 0.0151
iteration 2 residu = 0.00122
iteration 3 residu = 6.61e-05
iteration 4 residu = 9.23e-06
iteration 5 residu = 6.8e-07
iteration 6 residu = 1.37e-07
iteration 7 residu = 1.39e-08
iteration 8 residu = 3.24e-09
iteration 9 residu = 4.84e-10
iteration 10 residu = 1.07e-10
iteration 11 residu = 2.49e-11
iteration 12 residu = 4.67e-12
Numerical integration#
quad, dblquad, tplquad,… Fortran library QUADPACK.
import scipy.integrate as spi
x2=lambda x: x**2
4.**3/3 # int(x2) in [0,4]
21.333333333333332
spi.quad(x2,0.,4.)
(21.333333333333336, 2.368475785867001e-13)
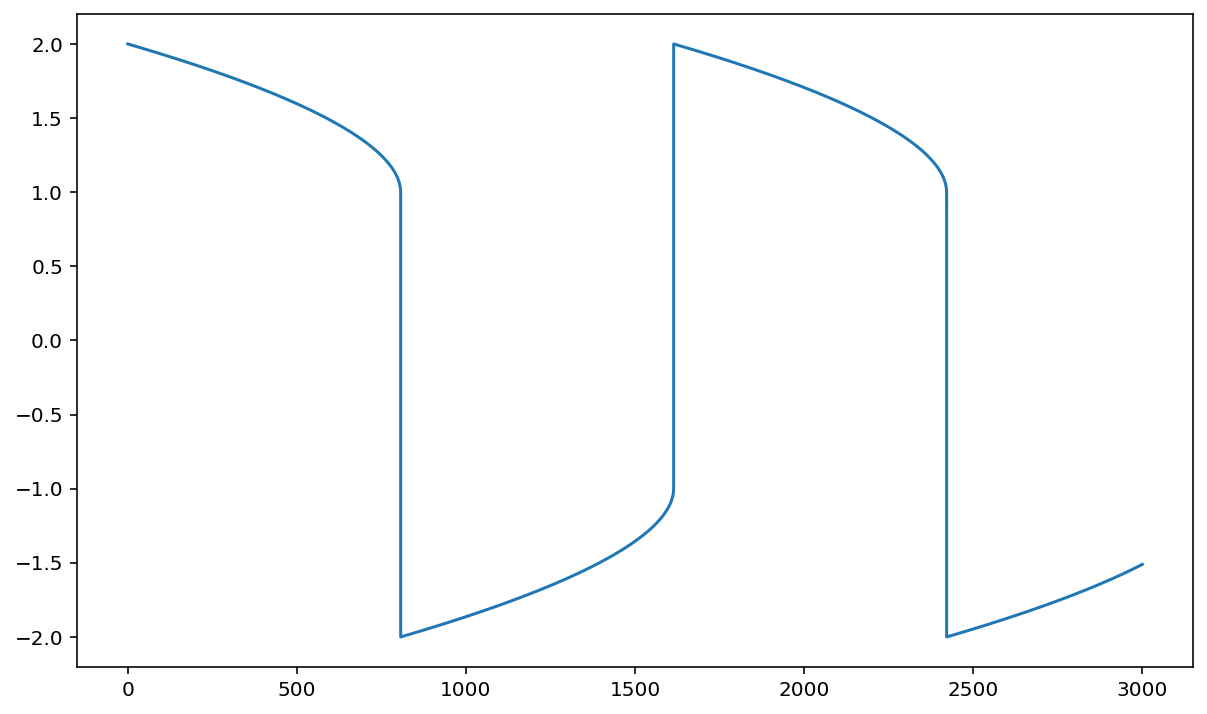
Scipy ODE solver#
It uses the Fortran ODEPACK library.
Van der Pol Oscillator#
\begin{align} y_1’(t) & = y_2(t), \nonumber \ y_2’(t) & = 1000(1 - y_1^2(t))y_2(t) - y_1(t) \nonumber \end{align} $\( with \)y_1(0) = 2 \( and \) y_2(0) = 0. $.
import numpy as np
import scipy.integrate as spi
def vdp1000(y,t):
dy=np.zeros(2)
dy[0]=y[1]
dy[1]=1000.*(1.-y[0]**2)*y[1]-y[0]
return dy
t0, tf =0, 3000
N = 300000
t, dt = np.linspace(t0,tf,N, retstep=True)
Exercise#
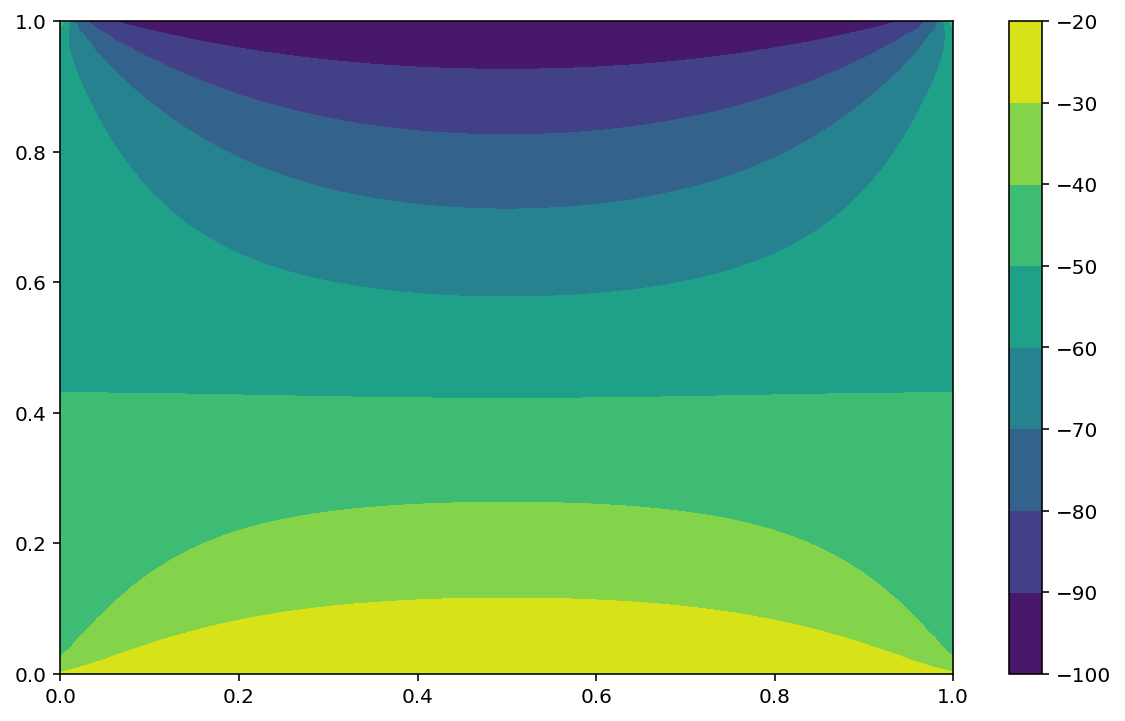
The following code solve the Laplace equation using a dense matrix.
Modified the code to use a sparse matrix
%matplotlib inline
%config InlineBackend.figure_format = "retina"
import numpy as np
import matplotlib.pyplot as plt
plt.rcParams['figure.figsize'] = (10,6)
# Boundary conditions
Tnorth, Tsouth, Twest, Teast = 100, 20, 50, 50
# Set meshgrid
n = 50
l = 1.0
h = l / (n-1)
X, Y = np.meshgrid(np.linspace(0,l,n), np.linspace(0,l,n))
T = np.zeros((n,n),dtype='d')
# Set Boundary condition
T[n-1:, :] = Tnorth / h**2
T[:1, :] = Tsouth / h**2
T[:, n-1:] = Teast / h**2
T[:, :1] = Twest / h**2
A = np.zeros((n*n,n*n),dtype='d')
nn = n*n
ii = 0
for j in range(n):
for i in range(n):
if j > 0:
jj = ii - n
A[ii,jj] = -1
if j < n-1:
jj = ii + n
A[ii,jj] = -1
if i > 0:
jj = ii - 1
A[ii,jj] = -1
if i < n-1:
jj = ii + 1
A[ii,jj] = -1
A[ii,ii] = 4
ii = ii+1
%%time
U = np.linalg.solve(A,np.ravel(h**2*T))
CPU times: user 662 ms, sys: 32 ms, total: 694 ms
Wall time: 405 ms
import scipy.sparse as spsp
import scipy.sparse.linalg as spspl
# Boundary conditions
Tnorth, Tsouth, Twest, Teast = 100, 20, 50, 50
# Set meshgrid
n = 50
l = 1.0
h = l / (n-1)
X, Y = np.meshgrid(np.linspace(0,l,n), np.linspace(0,l,n))
T = np.zeros((n,n),dtype='d')
# Set Boundary condition
T[n-1:, :] = Tnorth / h**2
T[ :1, :] = Tsouth / h**2
T[ :, n-1:] = Teast / h**2
T[ :, :1] = Twest / h**2
bdiag = -4 * np.eye(n)
bup = np.diag([1] * (n - 1), 1)
blow = np.diag([1] * (n - 1), -1)
block = bdiag + bup + blow
# Creat a list of n blocks
blist = [block] * n
S = spsp.block_diag(blist)
# Upper diagonal array offset by -n
upper = np.diag(np.ones(n * (n - 1)), n)
# Lower diagonal array offset by -n
lower = np.diag(np.ones(n * (n - 1)), -n)
S += upper + lower
%%time
U = sp.linalg.solve(S,np.ravel(h**2*T))
CPU times: user 822 ms, sys: 25.1 ms, total: 847 ms
Wall time: 529 ms